
1. Introduction
The whole big issue with foundational models is that it lacks real time inference skills. If a model was trained up to 2018, then it probably would not know anything about COVID. Now, there are a few ways around this:
- Fine Tuning: This is a fancy way of saying you are re-training the model with information different to what it was previously trained on. This is a perfectly good method in getting your model up to date, but it has some significant limitations like high cost, running out of parameters itself and the most important being loss of previous knowledge. Fine-tuning isn’t additive; this means that it may just replace old information with the new data it is being fed.
- Few Shot Prompting: If fine-tuning was too much of a hassle, prompt engineering is definitely easier. Just add the required context to the prompt itself and you’re good to go. This however also has very obvious limitations and it is impractical after a certain point (there’s no way you could add 22 million new Bio research papers into a prompt).
- RAG or Retrieval Augmented Generation is one of the most methods of updating the knowledge base of a pre-existing foundational model. [We’ll talk about RIG soon ;)]. So when the authors of this paper wanted to build a QA chatbot with an up to date knowledge base in a fast evolving and broad field like Biology, they used many techniques, chief among them being RAG.
2. Background
2.1 Retrieval-Augmented Generation (RAG)
Put simply RAG is like an “open book exam.”
-
All the newer (or relevant) content is chunked, embedded and stored in a Vector database. When the user now put forwards a query, that too is embedded and a cosine search is performed between the embedded query and embeddings present in the vector db to find the top K most relevant chunks of information.
a) Mathematical process:
- For each document vector (D) in the database and the query vector (Q):
- Calculate cosine similarity: cos(θ) = (D · Q) / (||D|| ||Q||) Where · is the dot product, and ||D|| and ||Q|| are vector magnitudes.
b) Interpretation:
- Cosine similarity measures the cosine of the angle between vectors.
- Range is from -1 to 1, where 1 means identical direction (most similar).
- In practice, for positive embeddings, the range is often 0 to 1.
c) Efficiency:
- Vector databases often use approximation algorithms (like HNSW or FAISS) to perform this similarity search efficiently on large datasets.
-
The prompt along with the k most relevant pieces of information to the initial query are bundled together and fed into the foundational model. [If you’re thinking this is technically a one shot prompt in the end, you are correct!] This increases the accuracy of results by a whole lot while neither being too expensive or risk losing previous context.
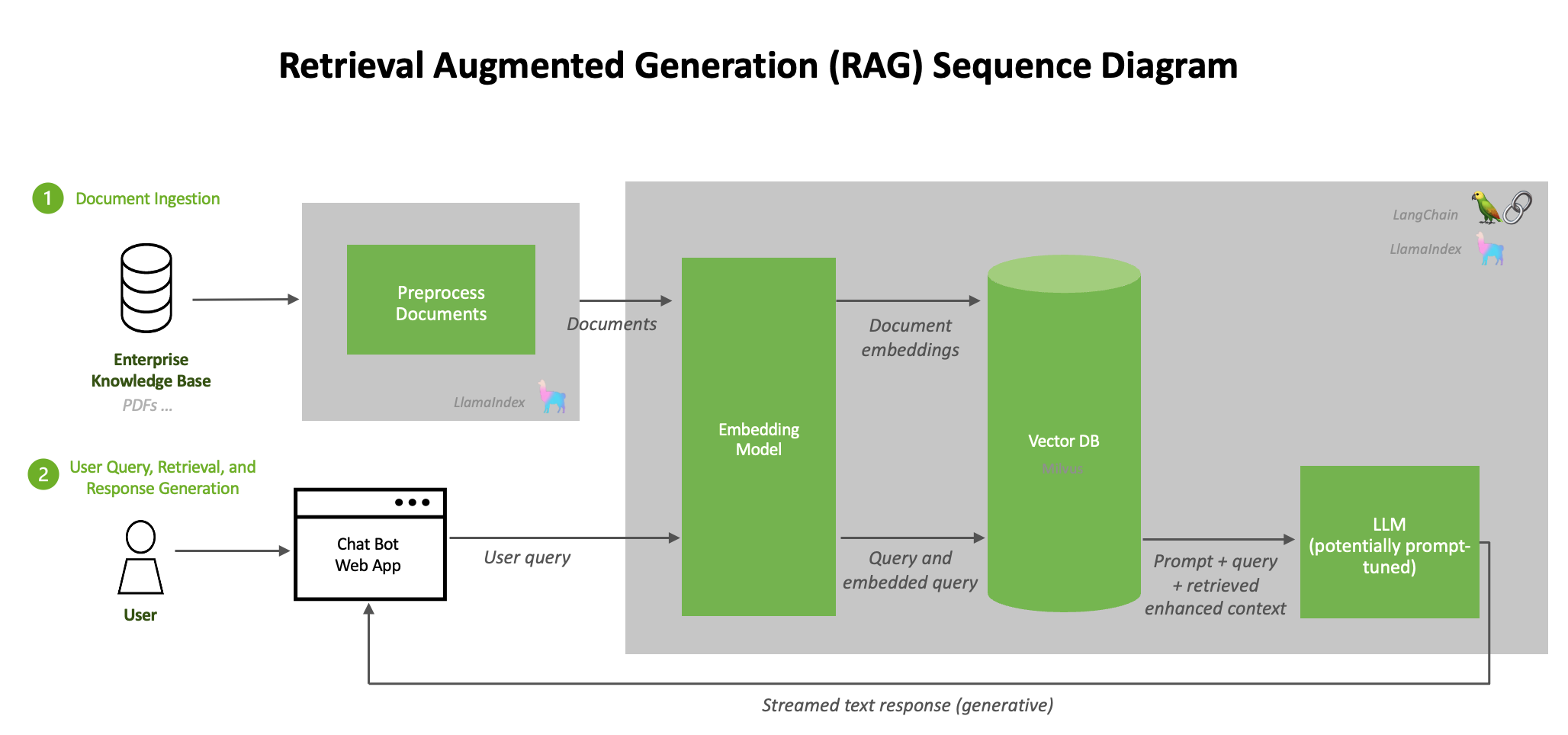
source: https://developer.nvidia.com/blog/rag-101-demystifying-retrieval-augmented-generation-pipelines/
Fun Fact: This exact technique is what Grok-0 used to get a 33B parameter model work as well as the LLaMA 2 (70B) parameter model, while also using half the training resources. This is also how Grok stays up to date with what’s going on on Twitter (sorry X).
2.2 The need for BioRAG - the most intensive biology question- reasoning system
- While fine tuned language models like bioBERT and sciBERT exist and are trained (fine tuned) with deep domain knowledge, this knowledge base is expensive to keep updated.
- On the other hand, RAG based systems, “often overlook the intricate complexities inherent in the domain knowledge of biology.”
- Hence, BioRAG was created to:
- Solve the scarcity of high-quality domain-specific corpora
- Capture the inherent complexity of biological knowledge systems
- Optimized continual updating of knowledge
3. BIORAG Architecture
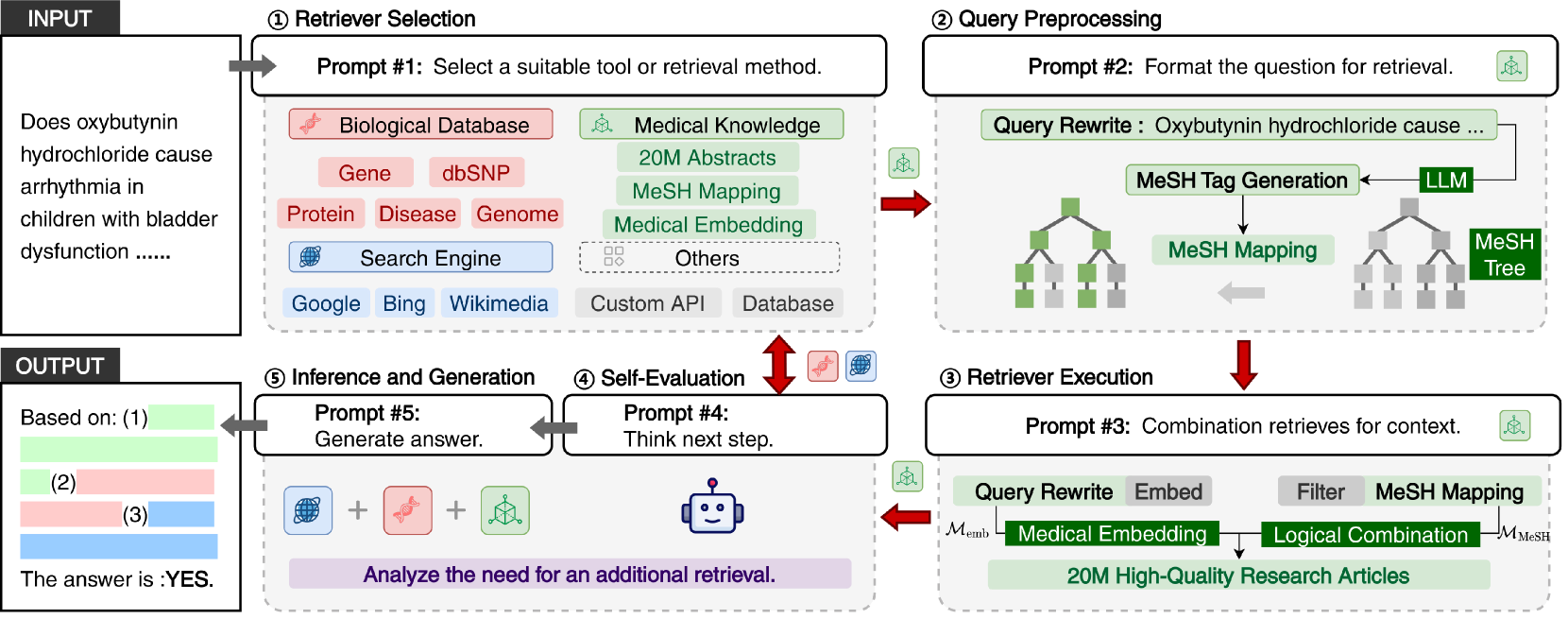
-
Retriever Selection: Choosing the Right Source
A prompt is used to analyze the query to determine which information sources are most relevant:
- Local PubMed Database: For established scientific knowledge
- Biological Data Hubs: For specific entity information (genes, proteins, etc.)
- Search Engines: For current research and general information
Query: "Latest treatment for BRCA1-positive breast cancer?" Source Selection: 1. Gene Database (for BRCA1 specifics) 2. PubMed (for treatment protocols) 3. Search Engines (for latest clinical trials) -
Query Pre-processing: Smart Query Understanding
This step transforms raw queries into structured formats that match each information source's requirements. It translates a casual question into a formal research inquiry.
-
Rewrites the query to suit the retrieval method chosen (by prompting).
-
MeSH Mapping: Maps to medical subject headings. Medical Subject Headings (MeSH) is like a hierarchical dictionary for biomedical terms. Think of it as a carefully organized tree where:
Diseases [C] ├── Cardiovascular Diseases [C14] │ ├── Heart Diseases [C14.280] │ │ ├── Heart Failure [C14.280.434] │ │ └── Arrhythmias [C14.280.067] └── Respiratory Tract Diseases [C08] ├── Lung Diseases [C08.381] └── Bronchial Diseases [C08.127]BIORAG uses a fine-tuned Llama model (M_MeSH) to predict relevant MeSH terms.
-
Finds related topics in knowledge hierarchy (based on MeSH Mapping).
-
-
Retriever Execution: Gathering Information
Retriever Execution aims to actually retrieve the relevant data from the knowledge base.
Query Input ↓ MeSH Filtering (if applicable) ↓ Vector Similarity Ranking (within the MeSH filtered results) ↓ Specialized Database Access (if needed) ↓ Self-Evaluation ↓ Additional Retrieval (if necessary)Multiple sources are not used simultaneously, but rather, sequentially. If the self evaluation mechanism deems it necessary for a different knowledge base to be used for additional information, we go back to the Retriever Selection step to choose a new knowledge base.
-
Self-Evaluation: Quality Control
This step acts as a quality control system, ensuring the retrieved information is sufficient and accurate. It's like having an expert reviewer checking if all necessary information has been gathered.
How It Works
- Completeness Check: Ensures all aspects of the query are addressed
- Relevance Assessment: Evaluates if information directly answers the query
- Confidence Scoring: Rates the reliability of information
- Gap Analysis: Identifies missing information
- New Retrieval: Goes back to a different knowledge base and retrieves missing information.
All this is done via a system prompt.
-
Inference and Generation: Creating the Answer
The final step puts together all gathered information into a coherent, accurate answer.
- Information Synthesis: Combines multiple sources
- Consistency Checking: Ensures no contradictions
- Evidence Ranking: Prioritizes most reliable information
- Response Formatting: Structures answer appropriately
3.1 Internal Biological Information Source
-
Out of the 37 million papers from NCBI’s PubMed Database, 22 million high quality abstracts were selected.
-
Unstructured was then used to further clean up the data. Unstructured is a tool for processing raw text data like :
- Text Extraction
- Removes HTML/XML tags
- Handles different file formats
- Preserves semantic content
- Cleaning Operations
- Removes special characters
- Standardizes whitespace
- Normalizes text encoding
- Handles tables and figures
- Content Structure Preservation
- Maintains paragraph breaks
- Preserves list structures
- Keeps important formatting
- Text Extraction
-
PubMedBERT is used as the foundational model as it is pre-trained on pubmed abstracts and full text articles.
-
CLIP was used to fine tune PubMedBERT. Lets talk about CLIP briefly.
-
CLIP (Contrastive Language-Image Pre-training)
This fundamentally refers to a technique of learning to match pairs of related content and using this contrastive learning to differentiate between related and unrelated pairs.
Goal: Learn to identify matching pairs while distinguishing from non-matching pairs Example Batch: ✓ (Image A, Text A) = Matching pair ✗ (Image A, Text B) = Non-matching pair ✗ (Image A, Text C) = Non-matching pair
Though the paper has not gone into how CLIP was implemented here, i assume they’ve created text-text pairs instead of the traditional image-text pairs.
An example to understand this:
Document: "Effects of Aspirin on Heart Disease" Positive Pairs: (Title, Abstract) (Introduction, Methods) (Related paper about aspirin effects) Negative Pairs: (Paper about genetics) (Paper about unrelated disease) (Random biomedical text) -
-
Using all this, finally a local vector database in constructed for effective query processing.
3.2 External Information Sources
- Apart from the Vector Database created, a way was also needed to keep on top of all the new information published daily without having to continually update the database.
- This was done using 2 instruments:
- Biological Data Hubs: Gene Database, dbSNP Database, Genome Database, Protein Database
- Real Time Search: Google & Bing, arXiv, Wikimedia, Crossref
- This real time search, along with the deep data knowledge from the fine tuned PubMedBERT makes BioRAG the most accurate QA system for biological tasks.
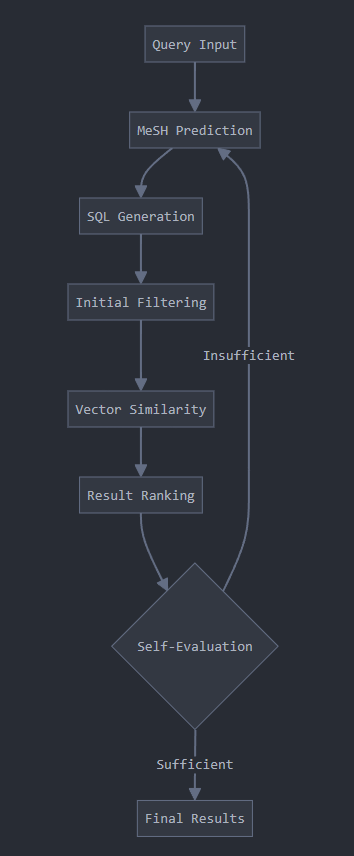
3.3 Self-evaluated Information Retriever
-
Internal Information Retrieve
-
As discussed above, MeSH is a system popularly used for cataloguing biomedical information. A model, M-MeSH was created by fine tuning a Llama3-8B model to predict MeSH from the input query.
-
Another really cool trick was using SQL for data retrieval. Since the query and the database have been broken down into MeSH, we can directly use SQL queries to match these, drastically speeding up inference.
-
Example: Scenario: Looking for papers about heart disease Papers Database: | Paper ID | Title | MeSH Terms | | 1 | Heart Study | Heart Disease, Treatment | | 2 | Brain Research | Brain, Neurons | | 3 | Heart Treatment | Heart Disease, Medicine | SQL Filter: SELECT * FROM papers WHERE mesh_terms LIKE '%Heart Disease%' Result: Returns papers 1 and 3
-
-
Self Evaluation Strategy
If the answer to the query is not adequate, another knowledge base, like web search for example will be used until the answer looks coherent and complete.
3.4 Final Comparisons
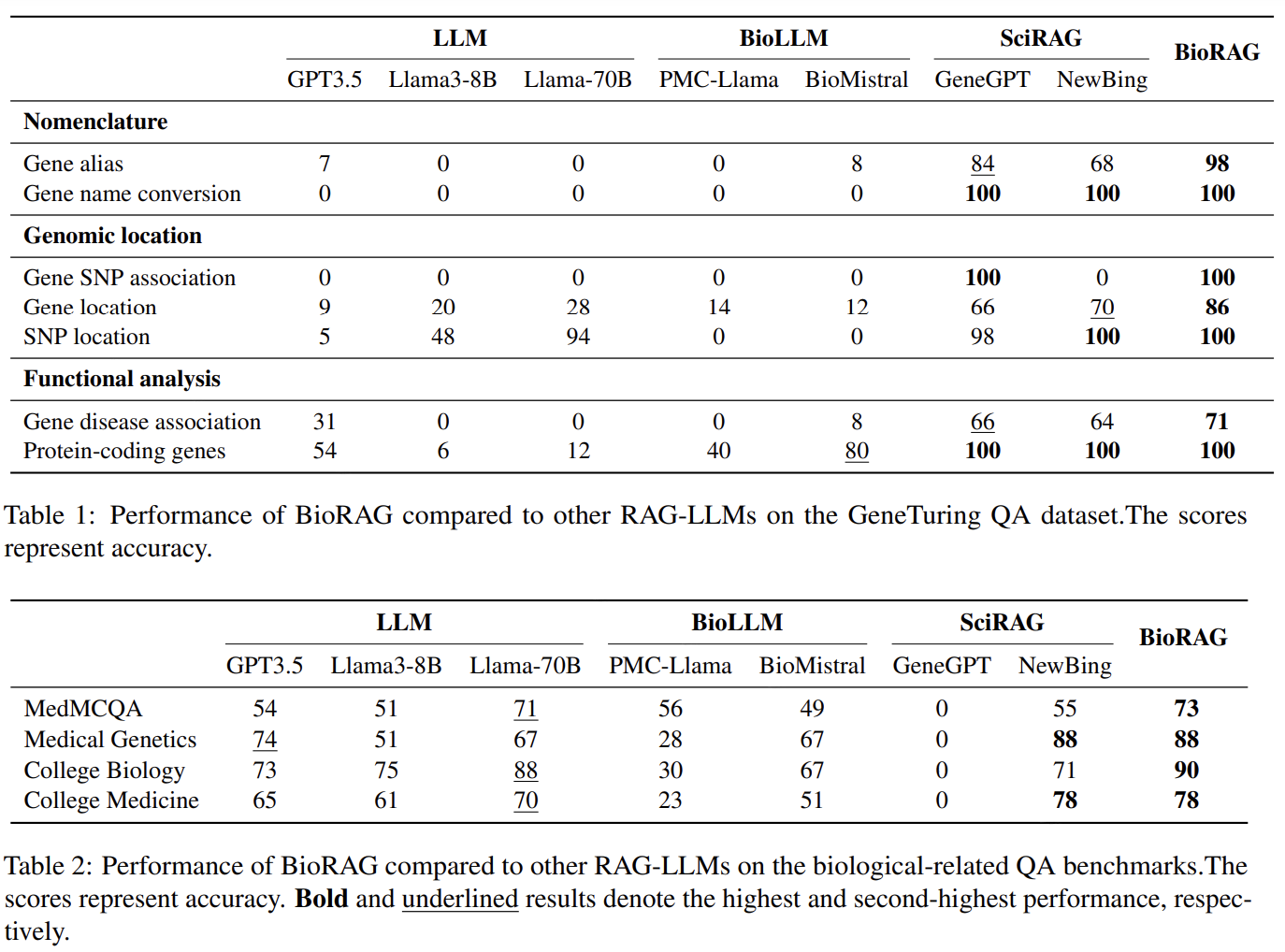
We're Done!
And that’s it! I covered this paper as it is one of the most complete information retrieval systems I have come across. I found it to be highly practical and usable in every field. Many small and smart tricks have been used to increase accuracy and efficiency. I intend to use these concepts to build a sort of paper implementation but add my own twist to it, do hit me up if you have any ideas!
Thanks for reading :)